This vignette will review the functionality for updating aesthetic
elements of VPC plots generated using vpc() with the
new_vpc_theme() function from the vpc
package.
plot_legend() is a helper plotting function that creates
a legend for plots generated using vpc(). These legends can
then be merged with the VPC plot into a single plot object using the
patchwork package.
Let’s get started. First, we will load the required packages.
options(scipen = 999, rmarkdown.html_vignette.check_title = FALSE)
library(pmxhelpr)
library(dplyr, warn.conflicts = FALSE)
library(ggplot2, warn.conflicts = FALSE)
library(vpc, warn.conflicts = FALSE)
library(mrgsolve, warn.conflicts = FALSE)
library(withr, warn.conflicts = FALSE)
library(patchwork, warn.conflicts = FALSE)Next, let’s load use the internal data and model objects from
pmxhelpr and df_mrgsim_replicate to run the
simulation.
data <- data_sad
model <- model_mread_load("model")
#> Building model_cpp ... done.
simout <- df_mrgsim_replicate(data = data, model = model,replicates = 100,
dv_var = "ODV",
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", DV = "DV"),
num_vars = c("CMT", "BLQ", "LLOQ", "EVID", "MDV", "DOSE", "FOOD"),
char_vars = c("PART"),
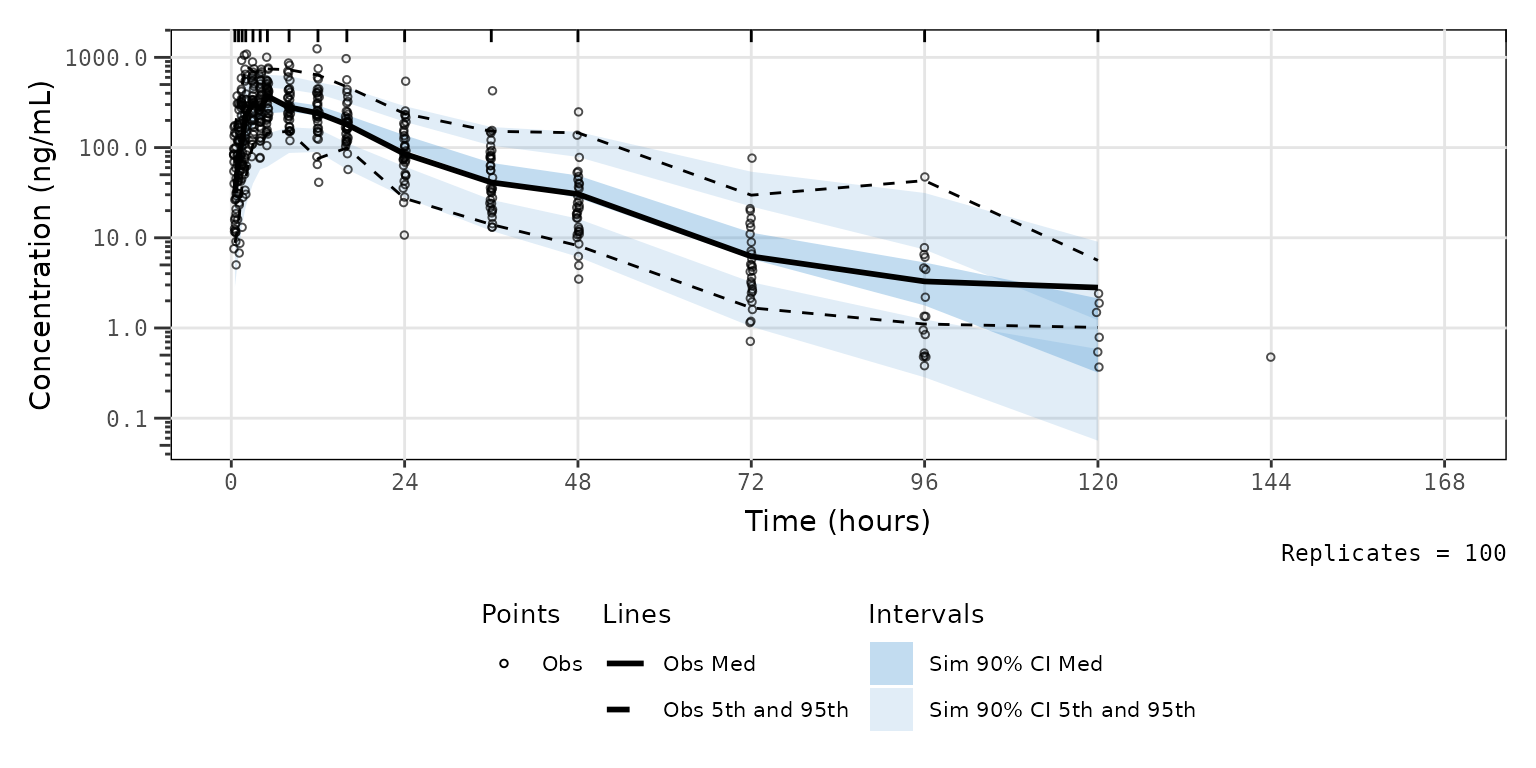
obsonly = TRUE)Now let’s plot all data together in a prediction-corrected VPC
(pcVPC). There is only single dose administration in this dataset, thus
we are able to pool across doses and food conditions with
prediction-correction. We will set min_bin_count = 5 to
avoid the simulated intervals extending to the final timepoint with only
a single observation.
vpc_pc <- plot_vpc_exactbins(
sim = simout,
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
min_bin_count = 5
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc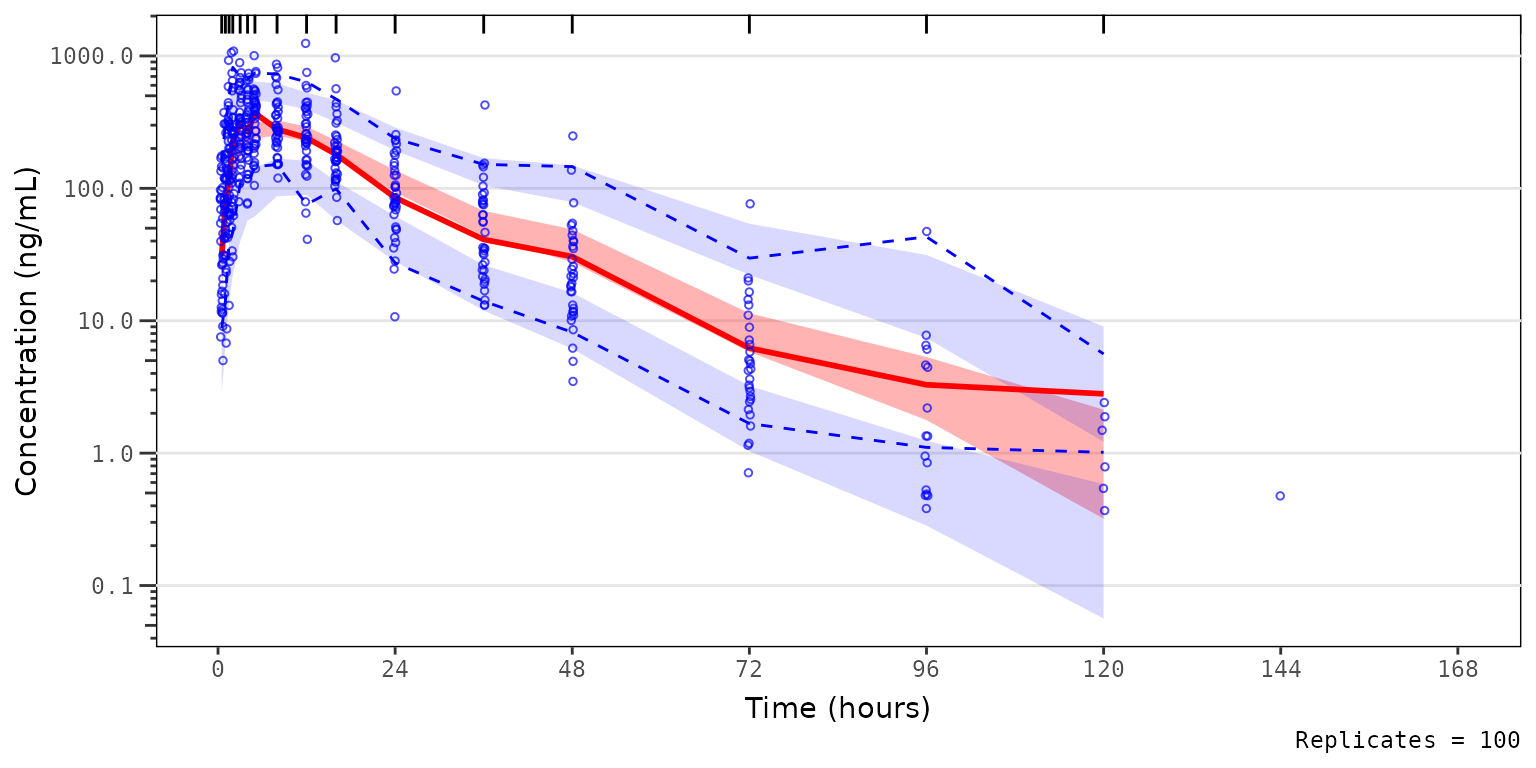
Plot Elements
Default Elements
The default elements shown in the plot are inherited from
vpc(). The shown argument can be provided to
plot_vpc_exactbins() and passed on to the show
argument of vpc().
The default is as follows:
shown_list <- list(obs_dv = TRUE, obs_ci = TRUE,
pi = FALSE, pi_as_area = FALSE, pi_ci = TRUE,
obs_median = TRUE, sim_median =FALSE, sim_median_ci = TRUE)The components of the list correspond to the following vpc plot elements:
- Observed points:
obs_dv - Observed quantiles:
obs_ci - Simulated inter-quantile range:
pi - Simulated inter-quantile area:
pi_as_area - Simulated Quantile CI:
pi_ci - Observed Median:
obs_median - Simulated Median:
sim_median - Simulated Median CI:
sim_median_ci
One or more elements to be updated from the defaults above can be
passed as a list to the argument shown. Any elements not
specified in shown will inherit the defaults.
Adjusting Elements
For example, we may want to visualize the 90% prediction interval (i.e., 5th to 95th percentiles) derived from the simulation, rather than the confidence intervals of the 5th and 95th percentiles independently. In this case, we want to also see how close the simulated median falls relative to the observed median.
Let’s see how this can be down using shown!
vpc_pc_pi <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", SIMDV = "SIMDV", OBSDV = "OBSDV"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
min_bin_count = 5,
shown = list(obs_ci = FALSE, pi_ci = FALSE, sim_median_ci = FALSE, sim_median = TRUE, pi_as_area = TRUE)
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_pi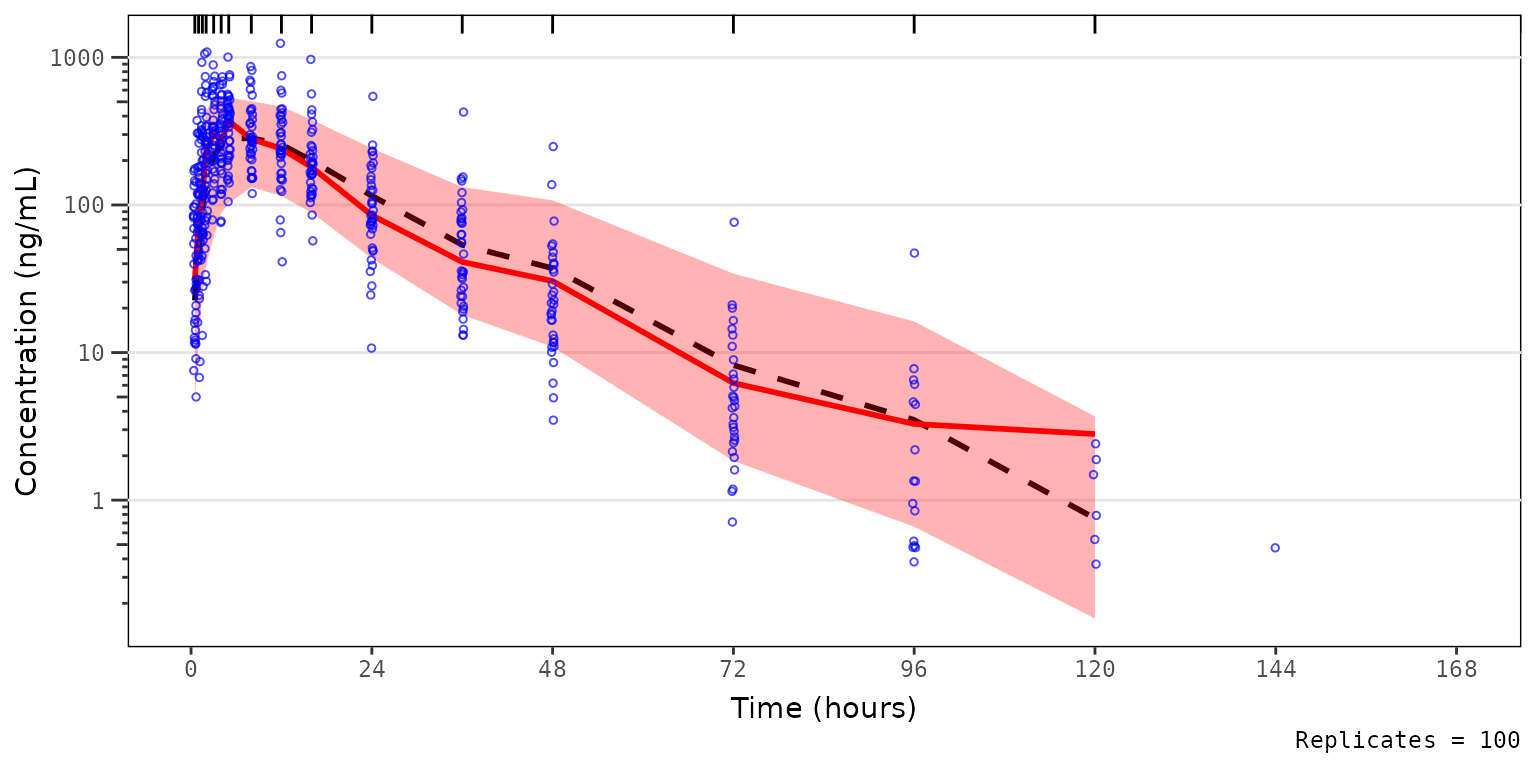
Now, let’s say we want to remove the observed data points from the plot above to better visualize the observed quantile lines relative to their corresponding simulated confidence intervals. We can do this as follows.
vpc_pc_noobs <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", SIMDV = "SIMDV", OBSDV = "OBSDV"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
min_bin_count = 5,
shown = list(obs_dv = FALSE)
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_noobs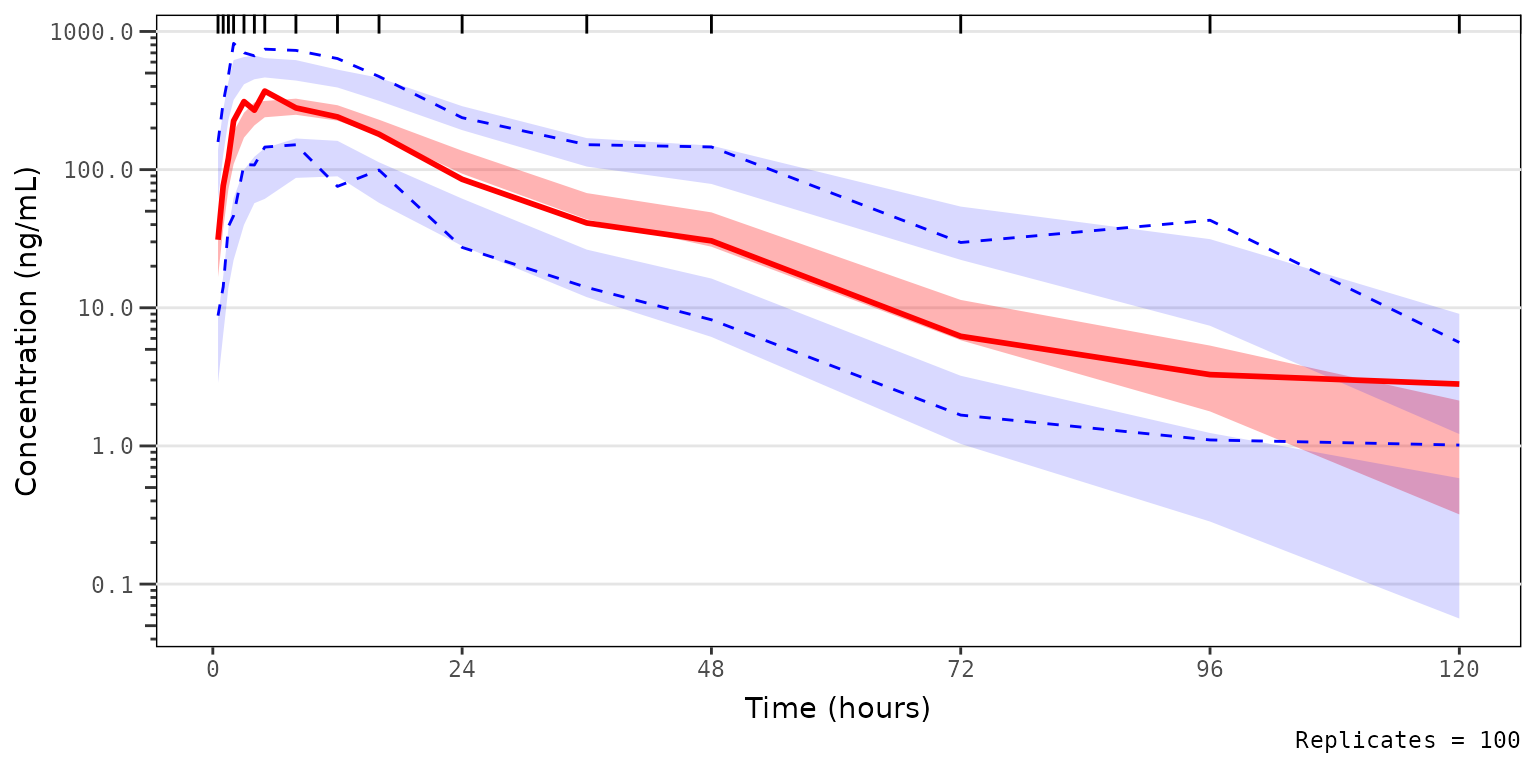
We could also take this one step further and only look at the median and the simulated confidence interval of the median, to closely interrogate central tendency. This is common for VPC strata which have few observations, leading to inadequate sample size to discriminate between the confidence intervals of the median and the extremes. This is common scenario when evaluating VPC plots stratified by individual study arms in early phase trials.
vpc_pc_noobs_medonly <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", SIMDV = "SIMDV", OBSDV = "OBSDV"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
min_bin_count = 5,
shown = list(obs_dv = FALSE, obs_ci = FALSE, pi_ci = FALSE)
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_noobs_medonly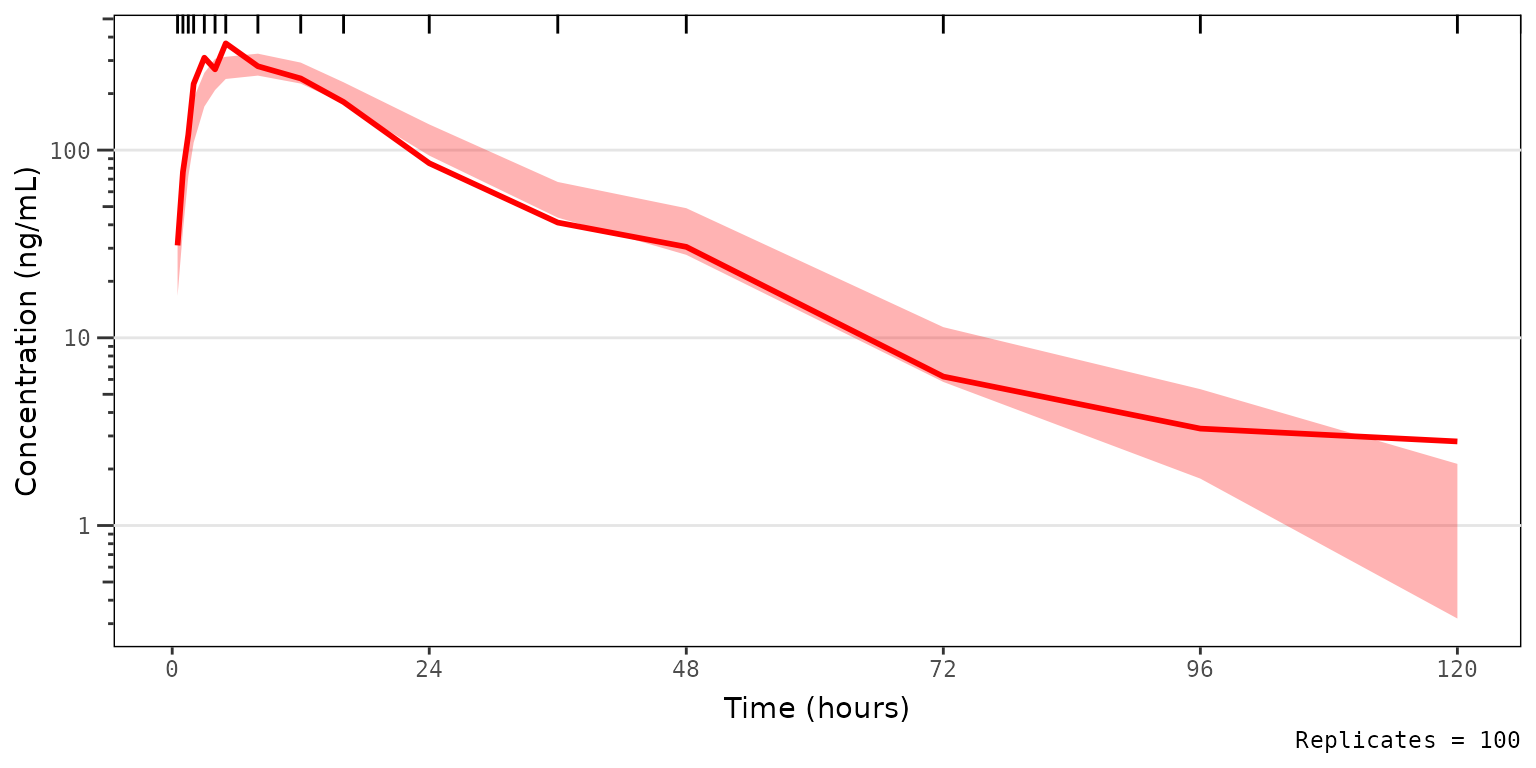
Plot Aesthetics
Default Aesthetics
Similarly, the default aesthetics for vpc plots in pmxhelpr use the
classical red-blue-green brewer color schema. The defaults for
pmxhelpr can be visualized by specifying the
pmxhlepr_vpc_theme() function with no arguments.
pmxhelpr_theme_list <- pmxhelpr_vpc_theme()
print(pmxhelpr_theme_list)
#> $obs_color
#> [1] "#0000FF"
#>
#> $obs_size
#> [1] 1
#>
#> $obs_median_color
#> [1] "#FF0000"
#>
#> $obs_median_linetype
#> [1] "solid"
#>
#> $obs_median_size
#> [1] 1
#>
#> $obs_alpha
#> [1] 0.7
#>
#> $obs_shape
#> [1] 1
#>
#> $obs_ci_color
#> [1] "#0000FF"
#>
#> $obs_ci_linetype
#> [1] "dashed"
#>
#> $obs_ci_fill
#> [1] "#80808033"
#>
#> $obs_ci_size
#> [1] 0.5
#>
#> $sim_pi_fill
#> [1] "#0000FF"
#>
#> $sim_pi_alpha
#> [1] 0.15
#>
#> $sim_pi_color
#> [1] "#000000"
#>
#> $sim_pi_linetype
#> [1] "dotted"
#>
#> $sim_pi_size
#> [1] 1
#>
#> $sim_median_fill
#> [1] "#FF0000"
#>
#> $sim_median_alpha
#> [1] 0.3
#>
#> $sim_median_color
#> [1] "#000000"
#>
#> $sim_median_linetype
#> [1] "dashed"
#>
#> $sim_median_size
#> [1] 1
#>
#> $bin_separators_color
#> [1] "#000000"
#>
#> $loq_color
#> [1] "#990000"
#>
#> attr(,"class")
#> [1] "vpc_theme"We can compare to the corresponding aesthetics from the
vpc package, which can be viewed by running
new_vpc_theme() with no arguments.
vpc_theme_list <- new_vpc_theme()
print(vpc_theme_list)
#> $obs_color
#> [1] "#000000"
#>
#> $obs_size
#> [1] 1
#>
#> $obs_median_color
#> [1] "#000000"
#>
#> $obs_median_linetype
#> [1] "solid"
#>
#> $obs_median_size
#> [1] 1
#>
#> $obs_alpha
#> [1] 0.7
#>
#> $obs_shape
#> [1] 1
#>
#> $obs_ci_color
#> [1] "#000000"
#>
#> $obs_ci_linetype
#> [1] "dashed"
#>
#> $obs_ci_fill
#> [1] "#80808033"
#>
#> $obs_ci_size
#> [1] 0.5
#>
#> $sim_pi_fill
#> [1] "#3388cc"
#>
#> $sim_pi_alpha
#> [1] 0.15
#>
#> $sim_pi_color
#> [1] "#000000"
#>
#> $sim_pi_linetype
#> [1] "dotted"
#>
#> $sim_pi_size
#> [1] 1
#>
#> $sim_median_fill
#> [1] "#3388cc"
#>
#> $sim_median_alpha
#> [1] 0.3
#>
#> $sim_median_color
#> [1] "#000000"
#>
#> $sim_median_linetype
#> [1] "dashed"
#>
#> $sim_median_size
#> [1] 1
#>
#> $bin_separators_color
#> [1] "#000000"
#>
#> $loq_color
#> [1] "#990000"
#>
#> attr(,"class")
#> [1] "vpc_theme"Adjusting Aesthetics
Now, suppose we want to change the default aesthetics of the VPC
plot, without changing what is being shown. This can be accomplished by
passing a named list of elements to update to the function
pmxhelpr_vpc_theme(). The named list object generated from
this function can then be passed to the theme argument in
vpc_plot_exactbins, which is also an alias for the
vpc_theme argument in vpc().
Let’s say we prefer a the blue-grey color scheme native to the
vpc package! We can reproduce this by passing
vpc::new_vpc_theme():
- to the update argument of
pmxhelpr_vpc_theme()
vpc_pc_vpctheme <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", SIMDV = "SIMDV", OBSDV = "OBSDV"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
theme = pmxhelpr_vpc_theme(update = vpc::new_vpc_theme()),
min_bin_count = 5
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_vpctheme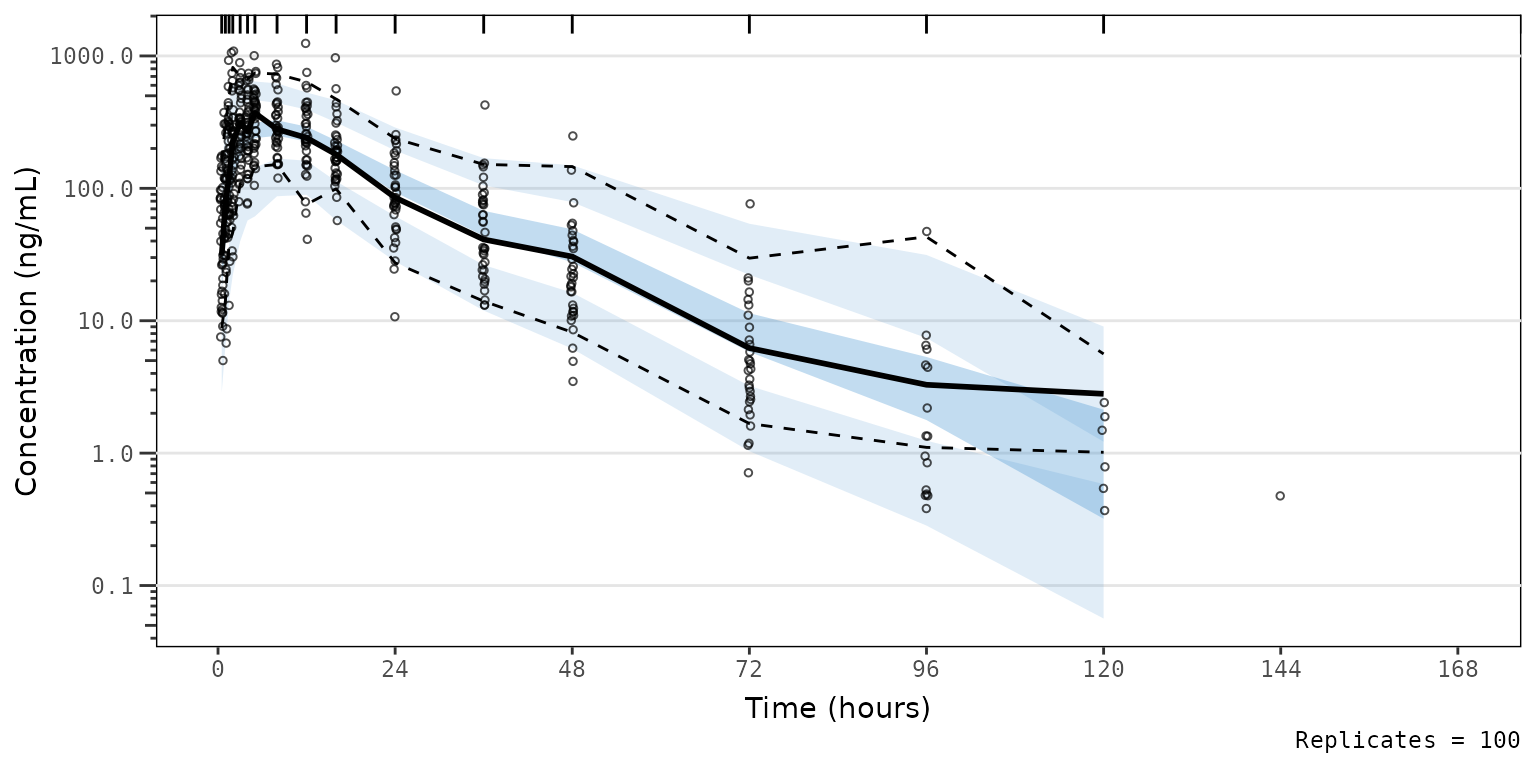 2) to the theme argument of
2) to the theme argument of plot_vpc_exactbins.
vpc_pc_vpctheme2 <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
output_vars = c(PRED = "PRED", IPRED = "IPRED", SIMDV = "SIMDV", OBSDV = "OBSDV"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
theme = vpc::new_vpc_theme(),
min_bin_count = 5
) +
scale_y_log10(guide = "axis_logticks")
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_vpctheme2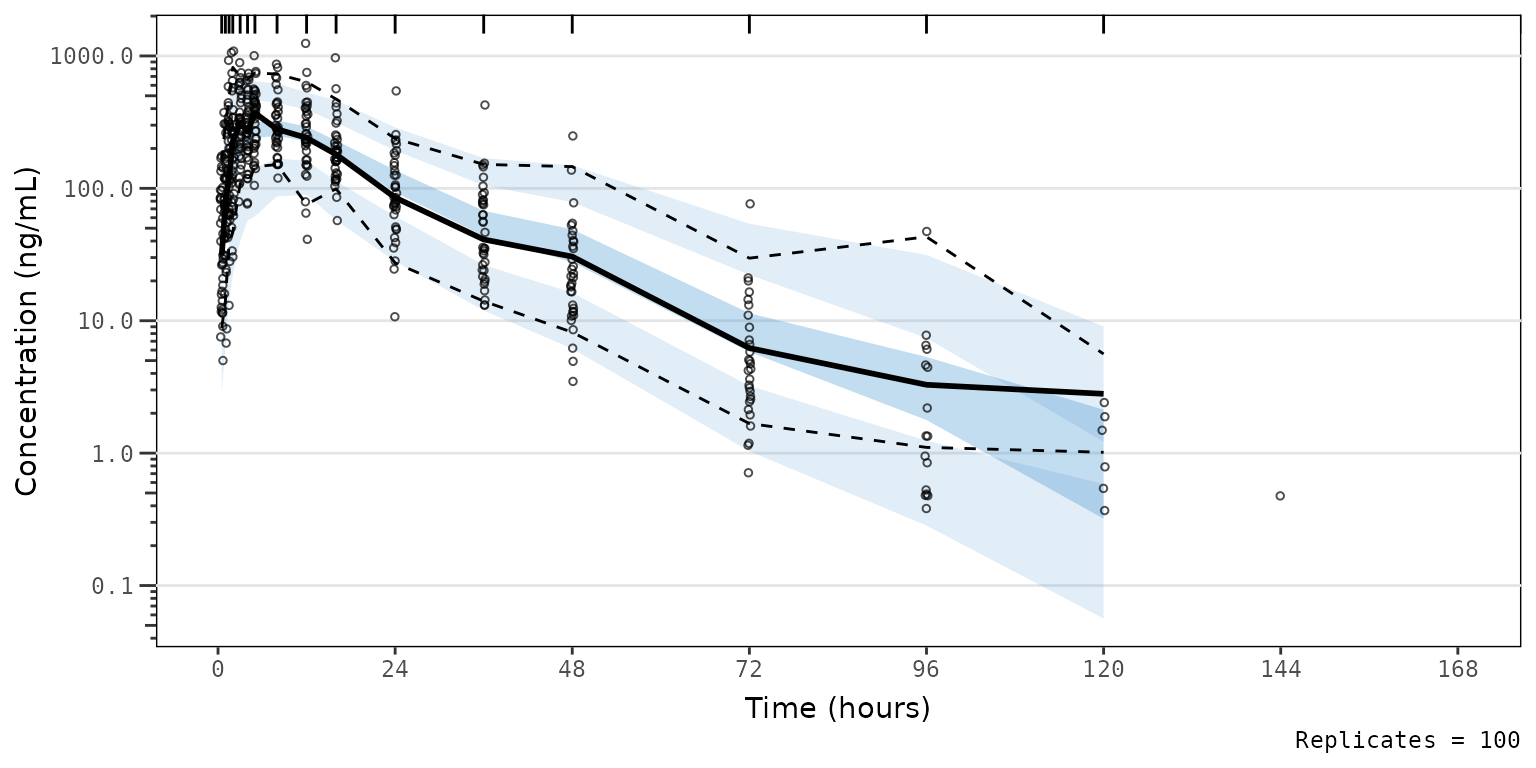
Adding Layers
Let’s say we would like to visualize the major x-axis grid lines to
help locate data corresponding to each sampling timepoint with our plot
using the vpc package aesthetic defaults. Conveniently,
plot_vpc_exactbins returns a ggplot2 plot
object, which we can modify by adding layers just like like any other
ggplot2 object.
vpc_pc_vpctheme_xgrid <- plot_vpc_exactbins(
sim = simout,
time_vars = c(TIME = "TIME", NTIME = "NTIME"),
xlab = "Time (hours)",
ylab = "Concentration (ng/mL)",
pcvpc = TRUE,
min_bin_count = 5,
theme = vpc::new_vpc_theme()
) +
scale_y_log10(guide = "axis_logticks") +
theme(panel.grid.major.x = element_line())
#> Joining with `by = join_by(NTIME, CMT)`
#> Joining with `by = join_by(NTIME, CMT)`
vpc_pc_vpctheme_xgrid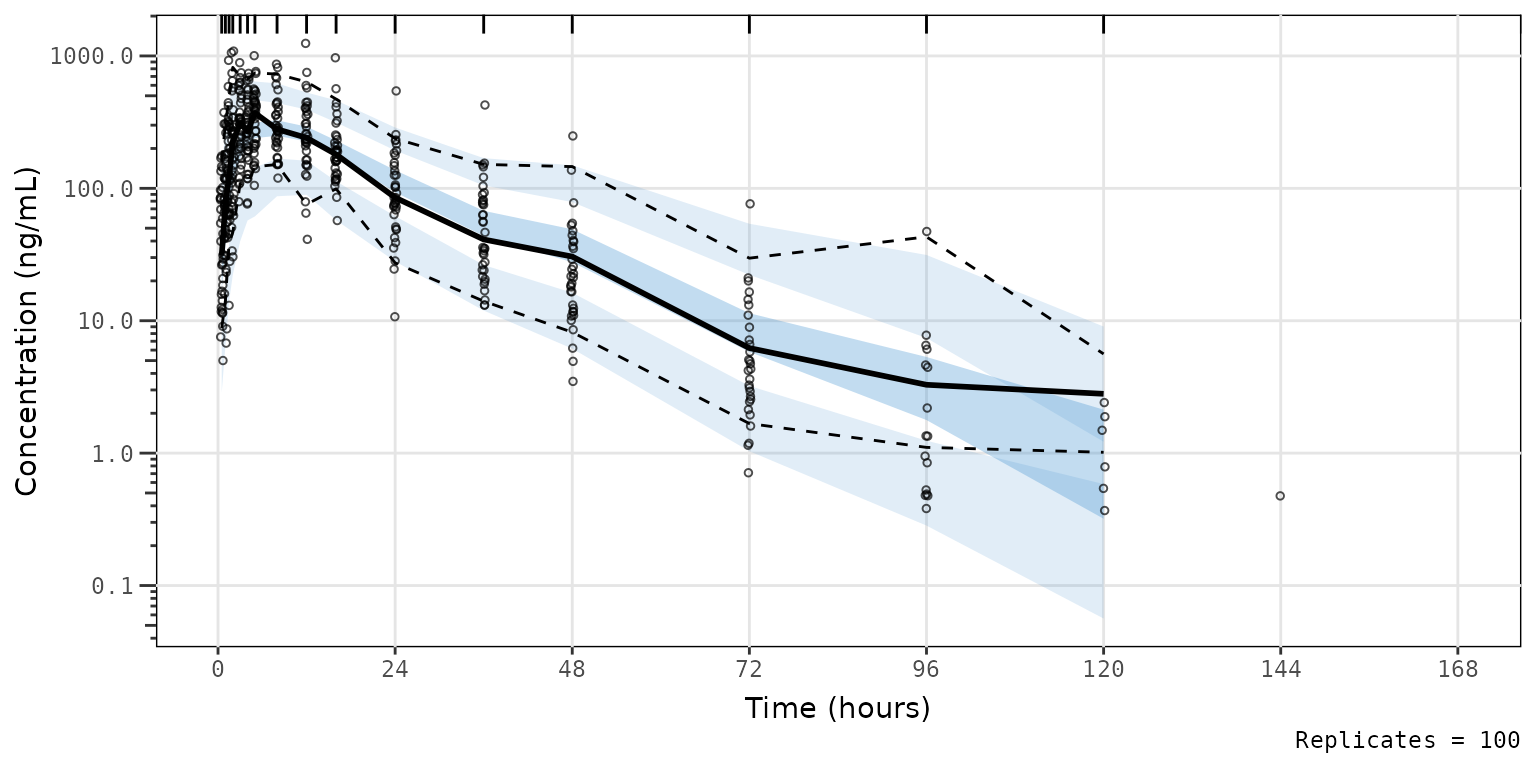
VPC Plot Legends
Defaults Legend
Okay, now we have gotten the plot aesthetics where we want them; however, there is one other element we may like to include in the figure to make it more easily interpreted in isolation - a legend.
pmxhelpr provides a useful helper function for this
purpose, plot_legend(). To obtain a legend for a plot using
default aesthetics, simply run plot_vpclegend() without any
arguments specified.
vpc_pc_legend <- plot_vpclegend()
vpc_pc_legend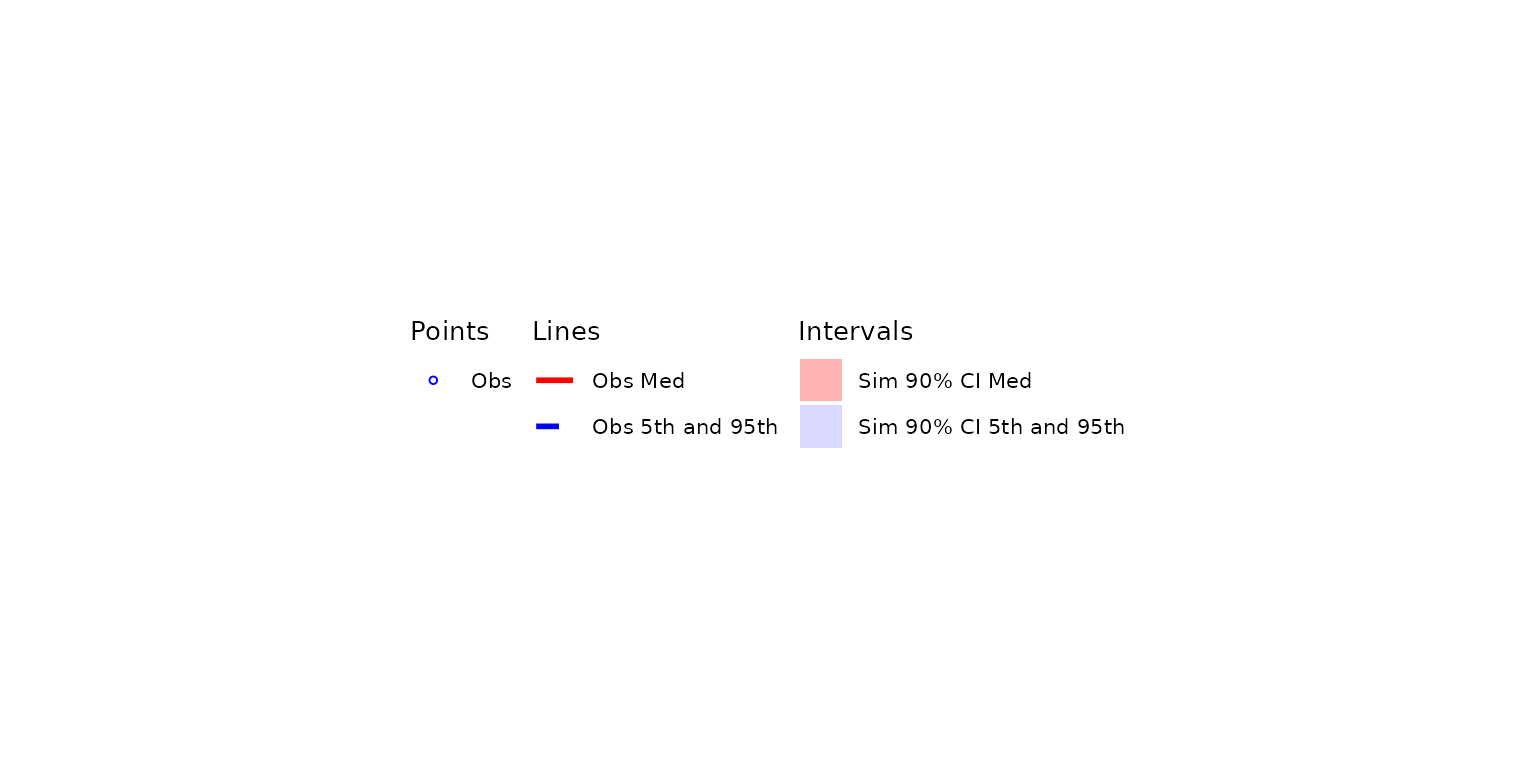 Now we have a
Now we have a ggplot object legend for our first plot!
To generate one for our second plot with updated aesthetics to match
the vpc package defaults, let’s pass the named list output
from vpc::new_vpc_theme() to the update
argument.
vpc_pc_new_legend <- plot_vpclegend(update = vpc::new_vpc_theme())
vpc_pc_new_legend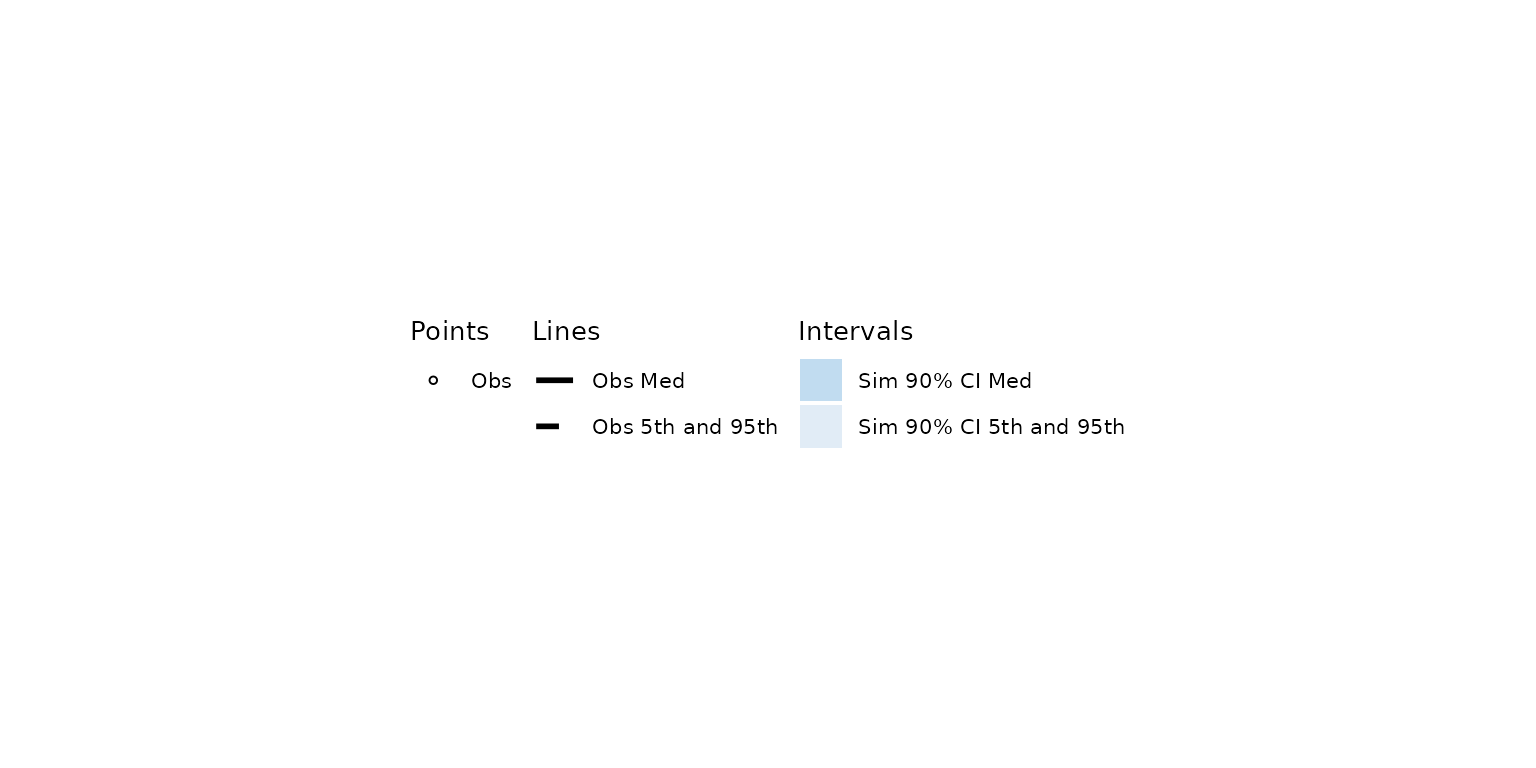
Okay, now that we have our legend plot objects, let’s combine them
with the VPC plot objects into a single plot object with the
patchwork package.
vpc_pc_wleg <- vpc_pc + vpc_pc_legend + plot_layout(heights = c(2.5,1))
vpc_pc_wleg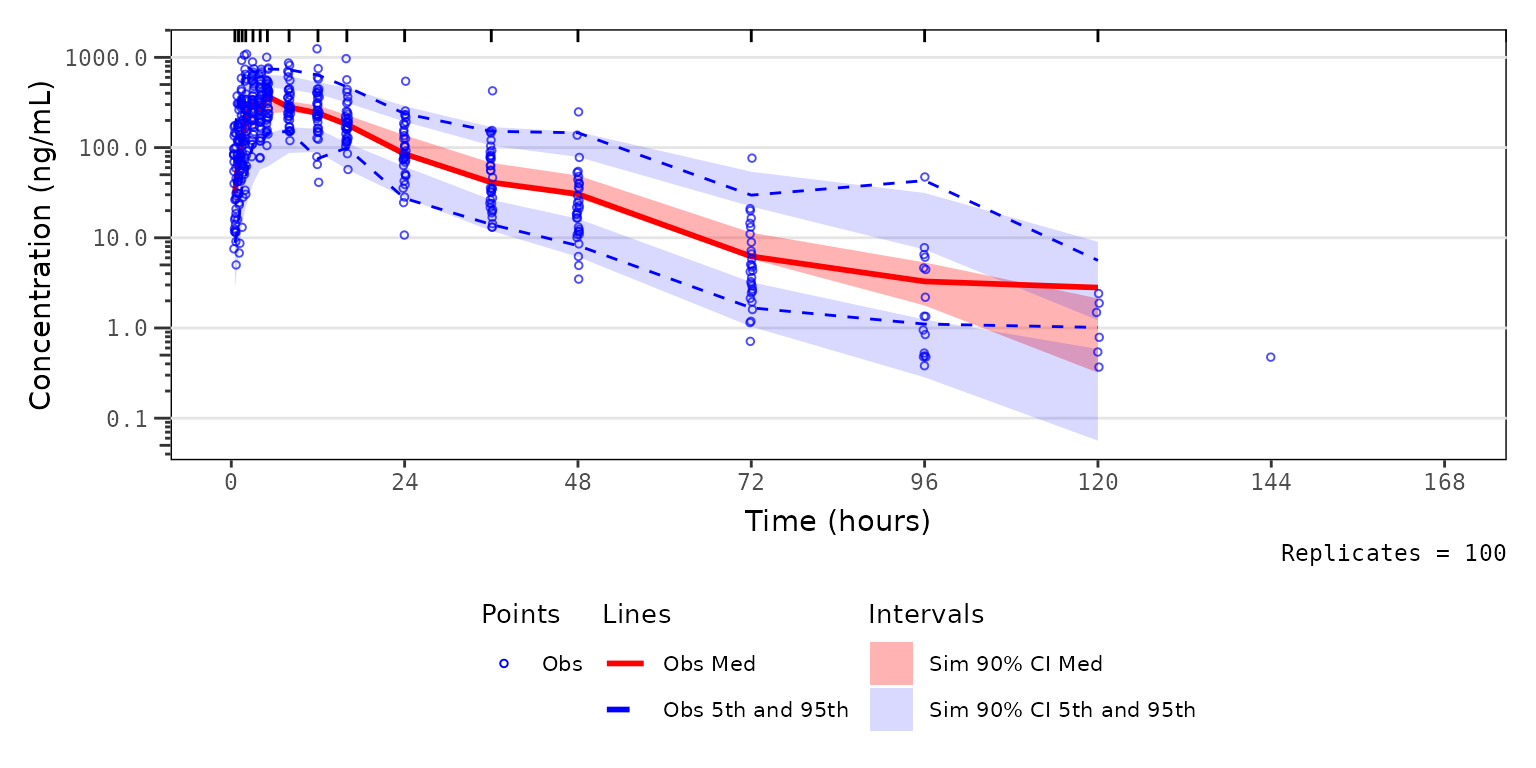
vpc_pc_new_wleg <- vpc_pc_vpctheme_xgrid + vpc_pc_new_legend + plot_layout(heights = c(2.5,1))
vpc_pc_new_wleg